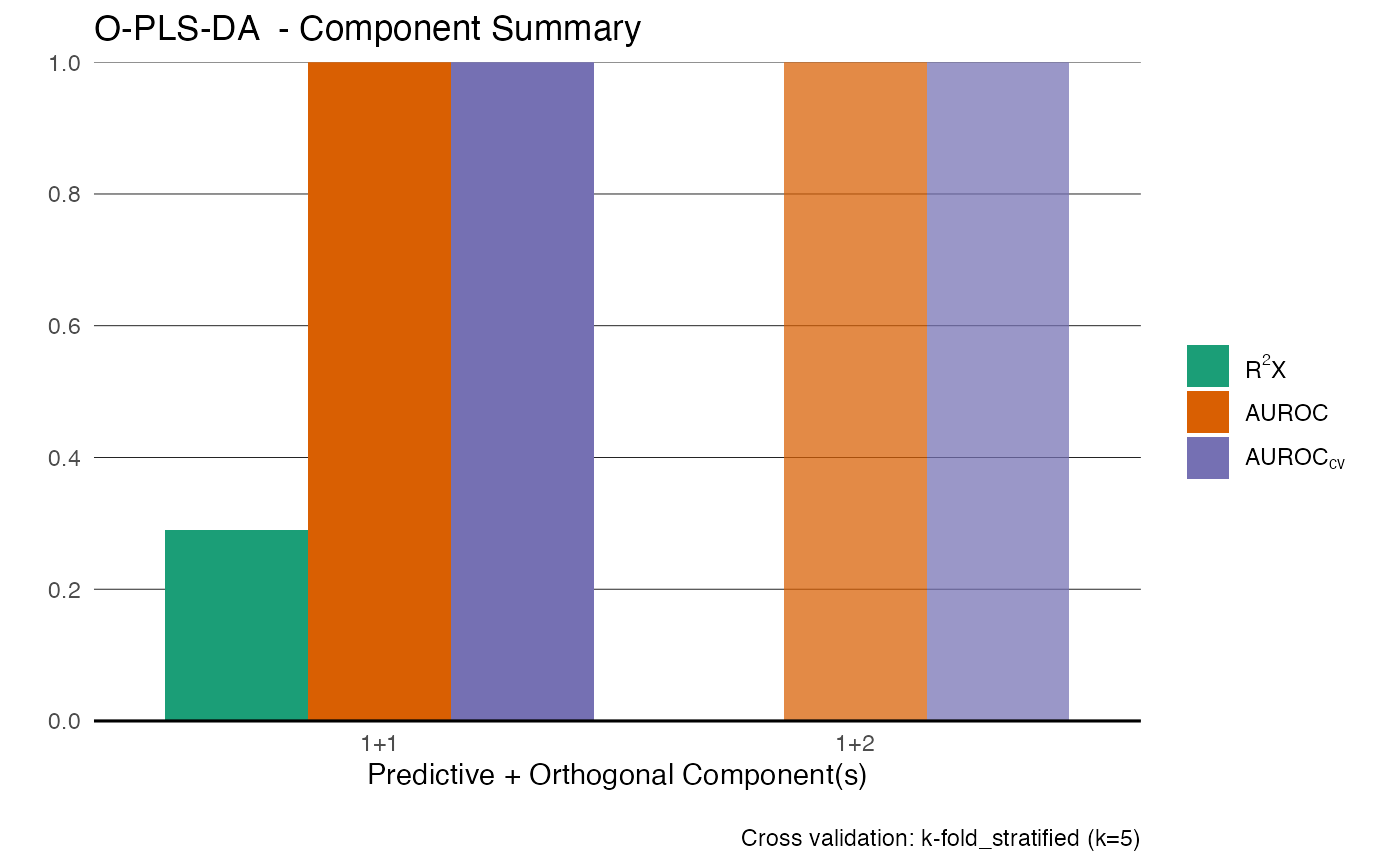
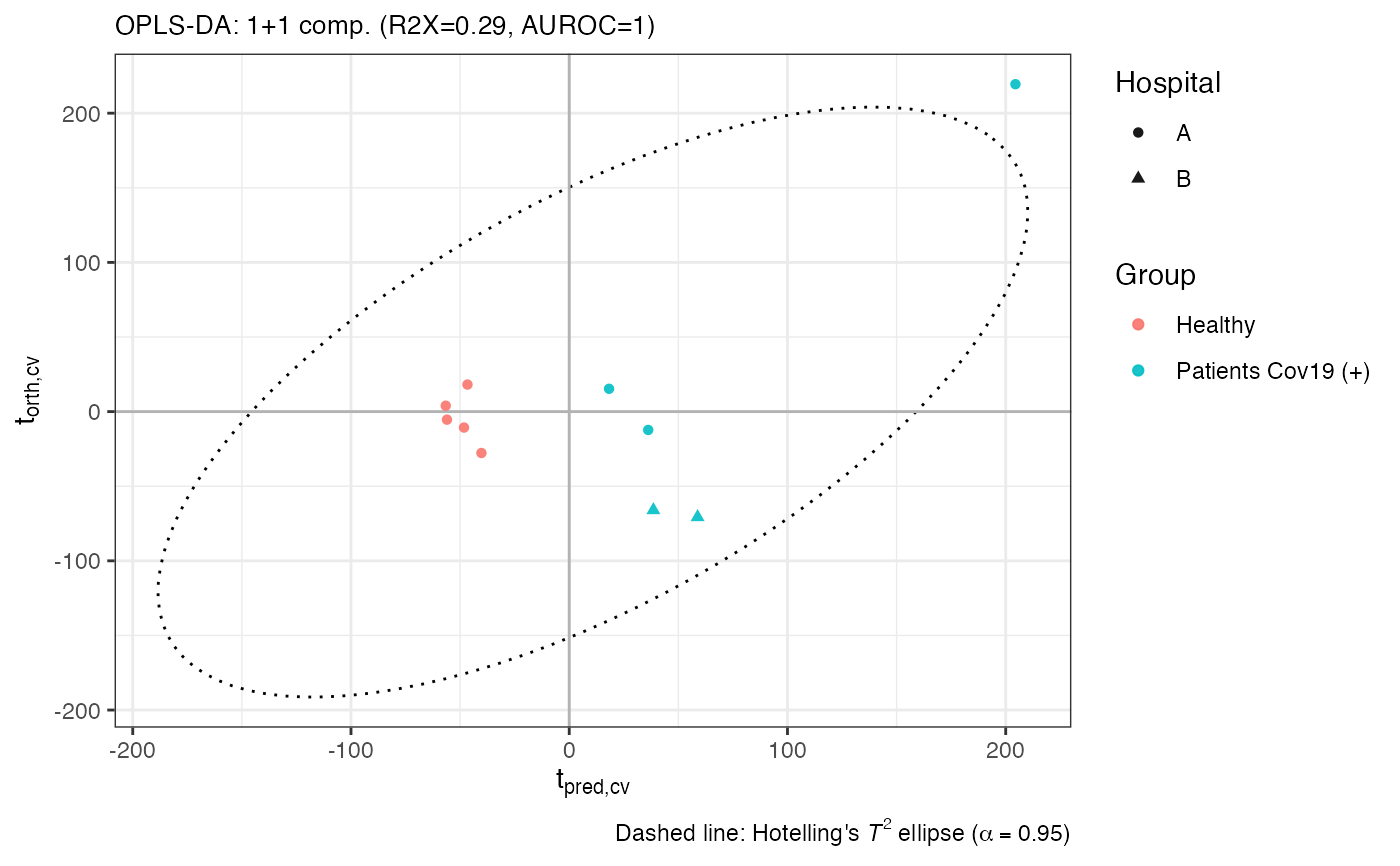
Generate score plots for PCA, PLS, or OPLS models with optional annotation, QC highlighting, and cross-validation scores.
Arguments
- obj
A fitted model object of class
PCA_metabom8,OPLS_metabom8,PLS_metabom8- pc
Numeric or character vector of length 2. Principal components or score components to plot. Defaults:
c(1,2)for PCA/PLS,c("1", "o1")for OPLS.- an
Optional list of up to 3 elements specifying annotation:
list(color, shape, label).- title
Optional character. Plot title.
- qc
Optional integer vector of row indices indicating QC samples to highlight.
- legend
Character. Legend position inside plot (default), or set to
NAto suppress.- cv
Logical. If
TRUE, cross-validated scores are shown when available (OPLS only).- ...
Additional arguments passed to
scale_colour_gradientn.
References
Trygg J., Wold S. (2002) Orthogonal projections to latent structures (O-PLS). Journal of Chemometrics, 16(3):119–128. Hotelling H. (1931) The generalization of Student’s ratio. Annals of Mathematical Statistics, 2:360–378.