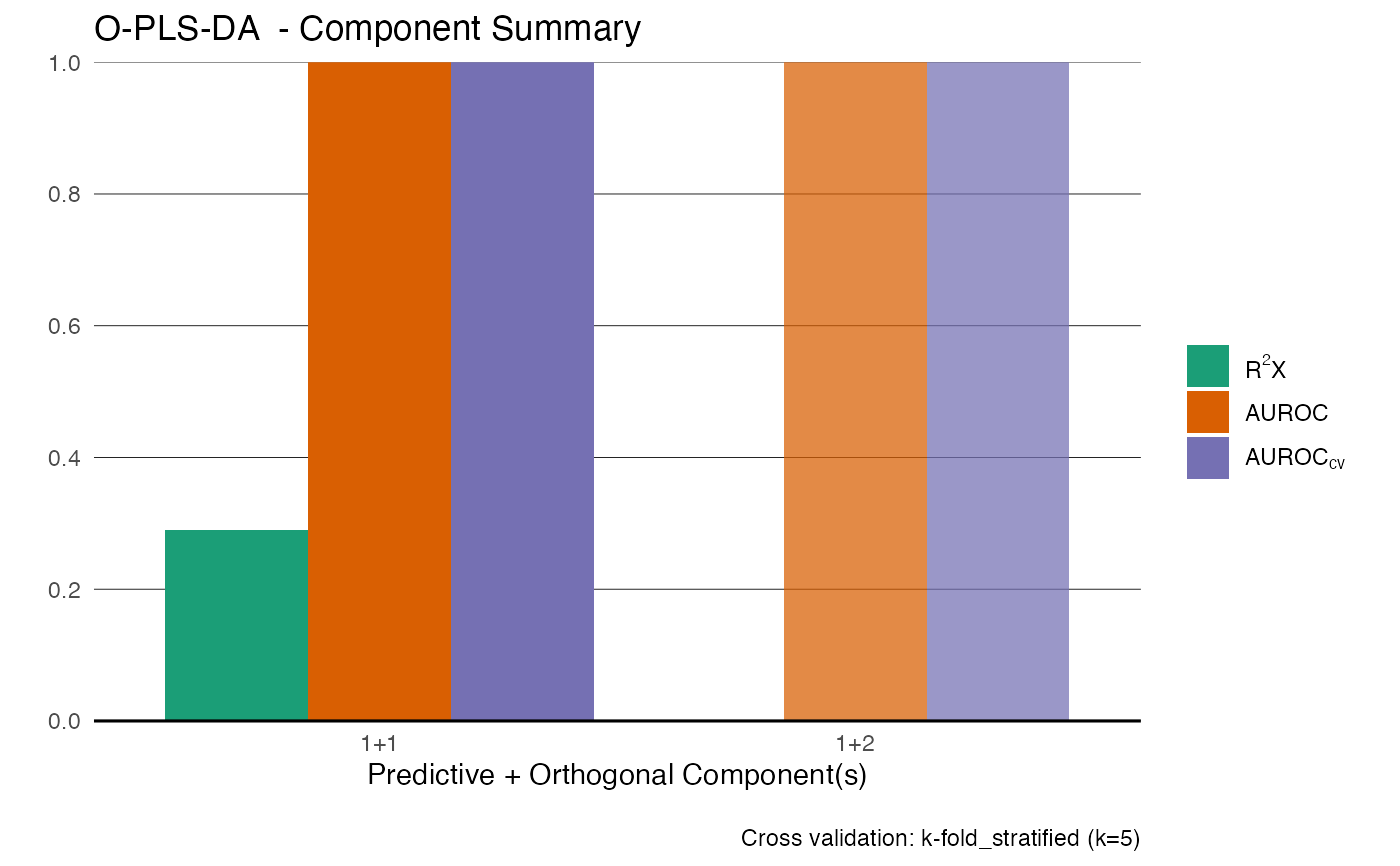
#' @title Orthogonal Partial Least Squares (O-PLS) #' #' @description #' Fits an Orthogonal Projections to Latent Structures (O-PLS) model for regression or classification. #' The number of orthogonal components is automatically selected using internal cross-validation. #' To avoid underfitting, components are added incrementally, while overfitting is prevented by requiring that each new component improves predictive performance beyond a defined threshold (ΔQ² / ΔAUROC > 0.05). This ensures the model captures relevant structure without modelling noise or irrelevant variation.
Source:R/opls.R
opls.Rd#' @title Orthogonal Partial Least Squares (O-PLS) #' #' @description #' Fits an Orthogonal Projections to Latent Structures (O-PLS) model for regression or classification. #' The number of orthogonal components is automatically selected using internal cross-validation. #' To avoid underfitting, components are added incrementally, while overfitting is prevented by requiring that each new component improves predictive performance beyond a defined threshold (ΔQ² / ΔAUROC > 0.05). This ensures the model captures relevant structure without modelling noise or irrelevant variation.
Usage
opls(
X,
Y,
center = TRUE,
scale = "UV",
cv = list(method = "k-fold_stratified", k = 7, split = 2/3),
maxPCo = 5,
plotting = TRUE
)Arguments
- X
Numeric matrix or data frame of predictors. Rows are samples; columns are features.
- Y
Response variable. Vector or matrix. For classification, should be a factor or class labels; for regression, a numeric vector or matrix.
- center
Logical. Should features be mean-centered? Default is TRUE.
- scale
Character. Scaling method. Supported: "None", "UV" (unit variance), or "Pareto".
- cv
Named list specifying cross-validation settings: method ("k-fold", "k-fold_stratified"), split (training set proportion), and k (number of folds).
- maxPCo
Integer. Maximum number of orthogonal components to consider. Default is 5.
- plotting
Logical. If TRUE, shows model summary plot.
References
Trygg J, Wold S. (2002). Orthogonal projections to latent structures (O-PLS). Journal of Chemometrics, 16(3), 119–128.