
MSbrowser is an R-based web-browser application for parameter fine-tuning of xcms peak picking algorithms.
The app is designed for fast and easy parameter testing to optimise xcms peak picking performance. Different visualisations allow to gain insight into the LC-MS data structure and the effects different peak picking parameter values have on peak picking performance.
MSbrowser is implemented in a user-friendly web-application framework that can be installed as a simple R package. Performance was tested with Google Chrome.
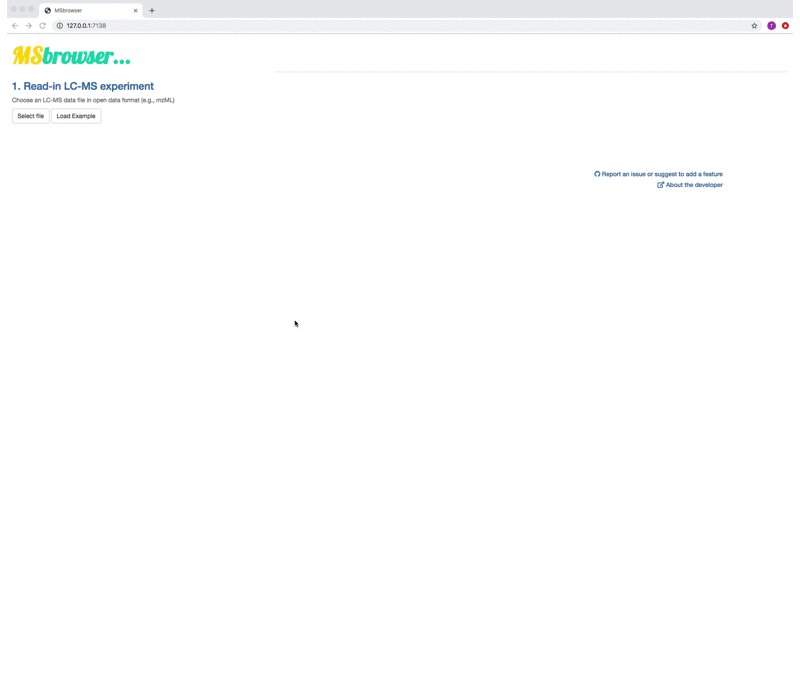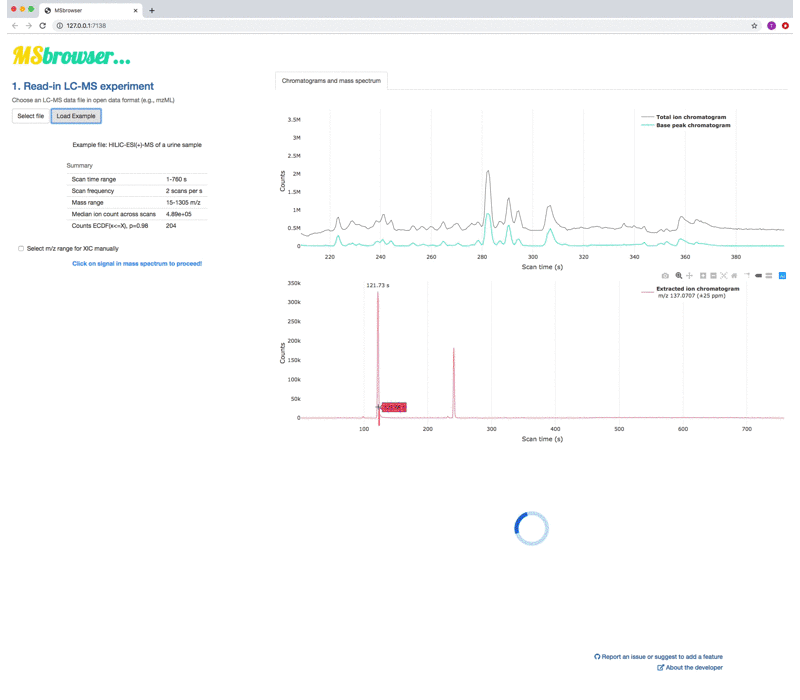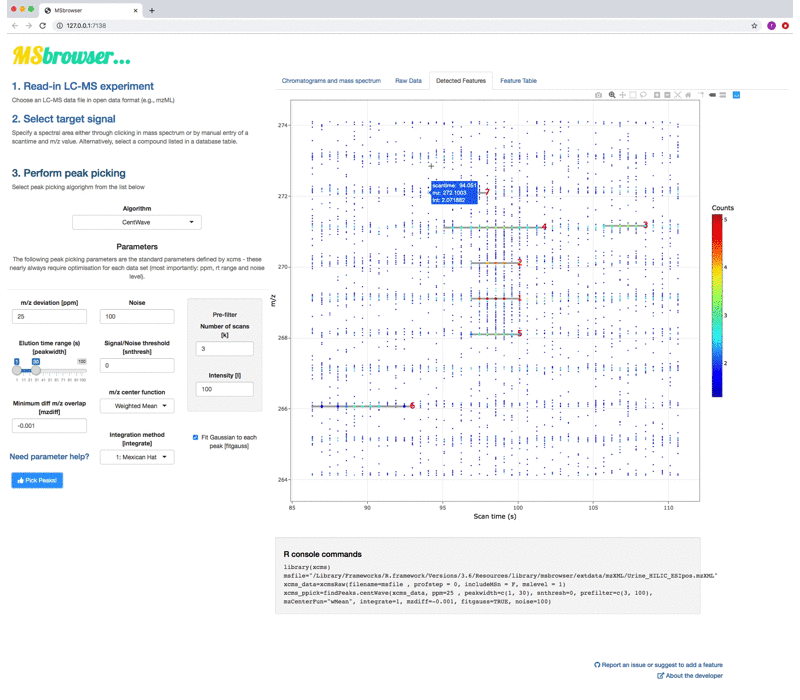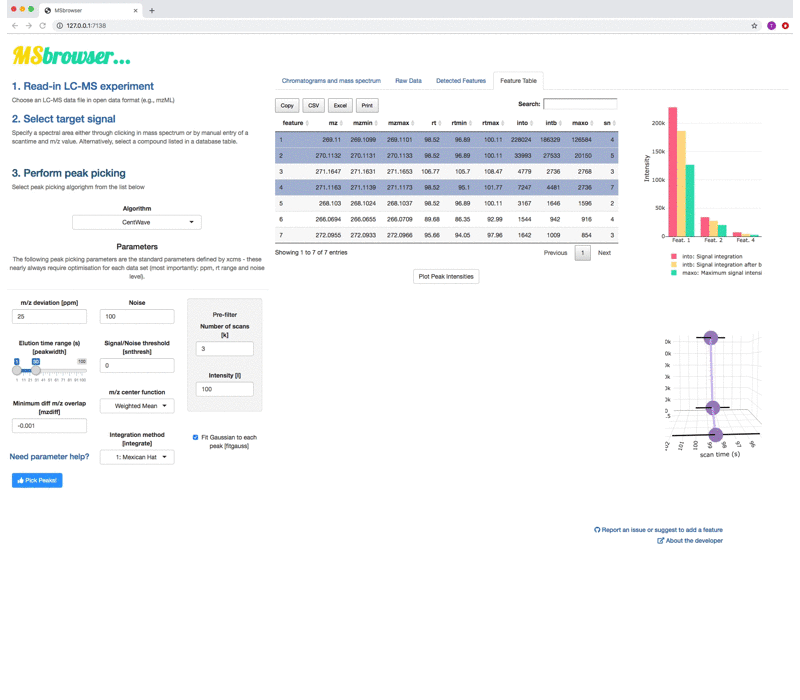
MSbrowser addresses the need to make peak picking of LC-MS data more transparent and reproducible across platforms! Key features of the app include:
- Generation of LC-MS experiment summary statistics
- User-interactive LC-MS data visualisatons that allow gaining insight into raw MS data structure
- Parameter testing and fine-tuning for xcms peak picking algorithms (centWave and matchFilter)
- Fast and interactive visualisations of xcms peak picking results
App installation and launch
The MSbrowser package is hosted on GitHub and is build with xcms version 3.6 (or higher) to perform peak picking. The following R code can be used to install necessary R dependencies on your computer.
# install dependencies from CRAN (devtools and BiocManager)
deps=c('devtools', 'BiocManager')
id=deps %in% installed.packages()[,1]
if(any(!id)) install.packages(deps[id])
# install xcms
if(!requireNamespace("xcms", versionCheck=list(op = ">=", version = "3.6"), quietly = T)){BiocManager::install('xcms')}Now you’re ready to go for the installation of MSbrowser:
# install MSbroswer
devtools::install_github('tkimhofer/msbrowser')If prompted by the command line, perform necessary package updates.
MSbrowser can be launched with the following R-terminal command:
msbrowser::startApp()A new web-browser window opens with the MSbrowser user interface.
Documentation
MSbrowser has an intuitive workflow, with help text placed in the user interface. Basic app functionalities and an illustration of the general workflow for fine-tuning of xcms peak picking parameters can be found in this video tutorial.
A Guide for centWave peak picking parameters can be found under the centWave Resources option in the page menu above.
MSbrowser can read MS files in open data formats mzML, mzXML, CDF, netCDF; file conversions from vendor format can be performed with open source softwares available either as a standalone (e.g., ProteoWizard) or online tool (e.g., GNPS).
Example Data
The application can be tested using human urine experiment data acquired with HILIC-ESI(+)-Q-TOF-MS. This data are available in the lcmsData package which can be installed with the following R code:
devtools::install_github('tkimhofer/lcmsData')Feedback
Got questions or suggestions? Log an issue on GitHub (requires login) or drop me an email!
